MARVEL – Metagenomic Analysis and Retrieval of Viral Elements
MARVEL is a tool for recovery of complete phage genomes from whole community shotgun metagenomic sequencing data.
A complete article describing MARVEL was published in Frontiers in Genetics.
Please, cite:
Amgarten, D. E., Braga, L. P. P., Da Silva, A. M., & Setubal, J. C. (2018). MARVEL, a Tool for Prediction of Bacteriophage Sequences in Metagenomic Bins. Frontiers in genetics, 9, 304.
MARVEL is publicly available at GitHub, along with installation instructions and usage examples:
GitHub repository
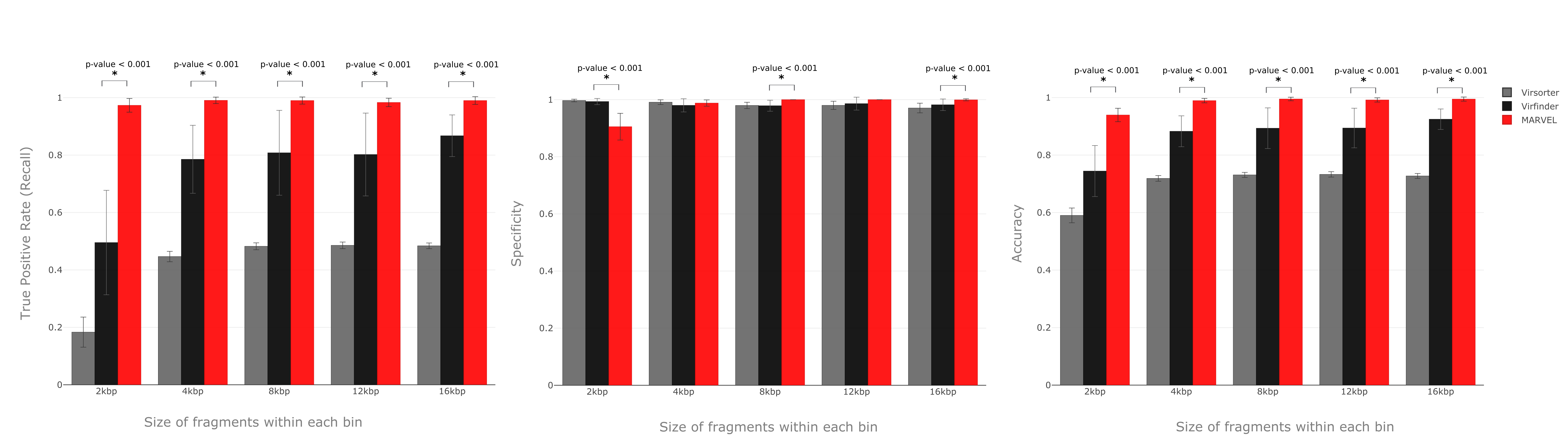
We implemented a random forest algorithm as MARVEL’s prediction core. Validation tests with mocking datasets from NCBI refseq database show accuracies, true positive rates and specificity all higher than 99% for bins composed of contigs 4 kbp or longer.
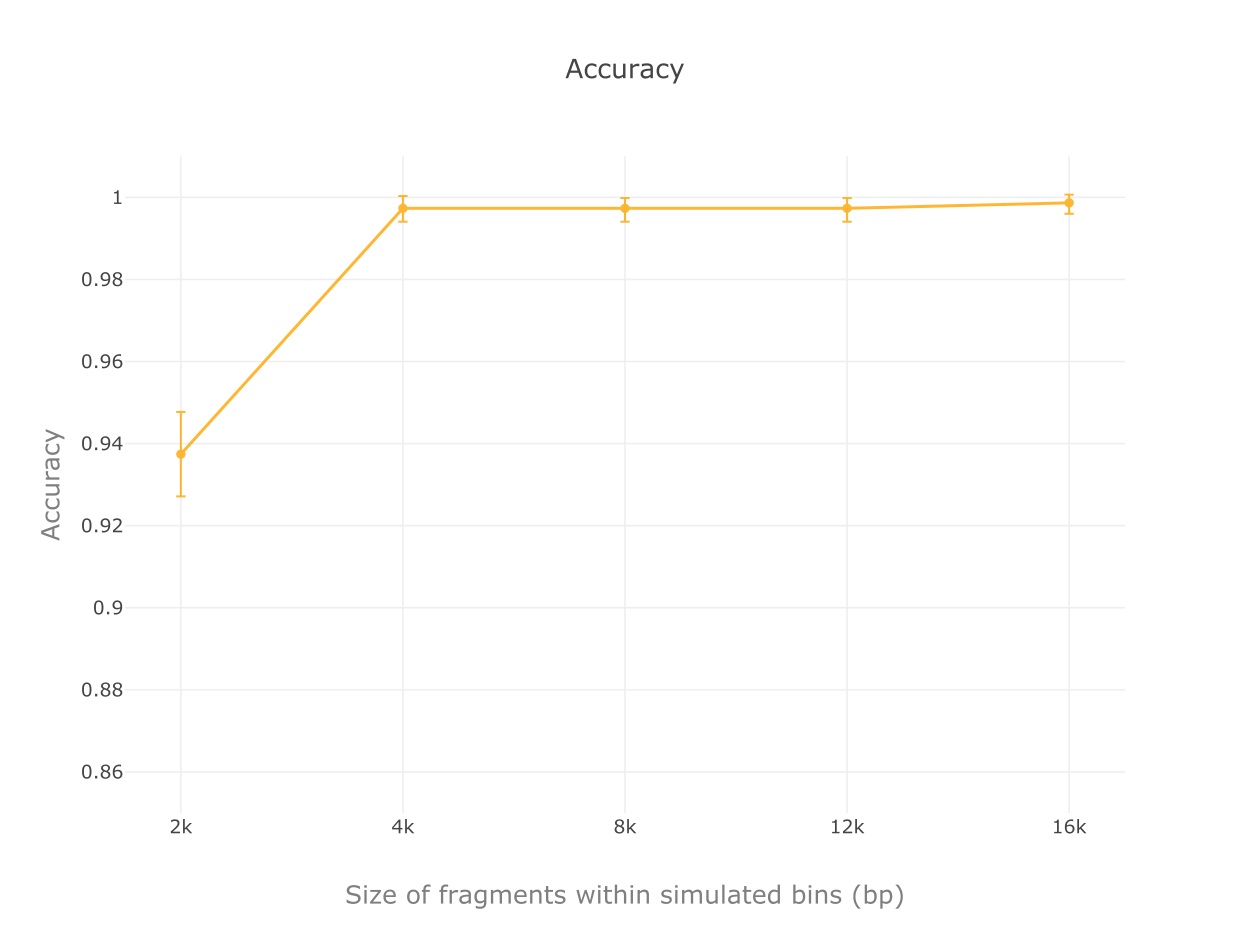
Accuracy results in tests for different contig sizes.
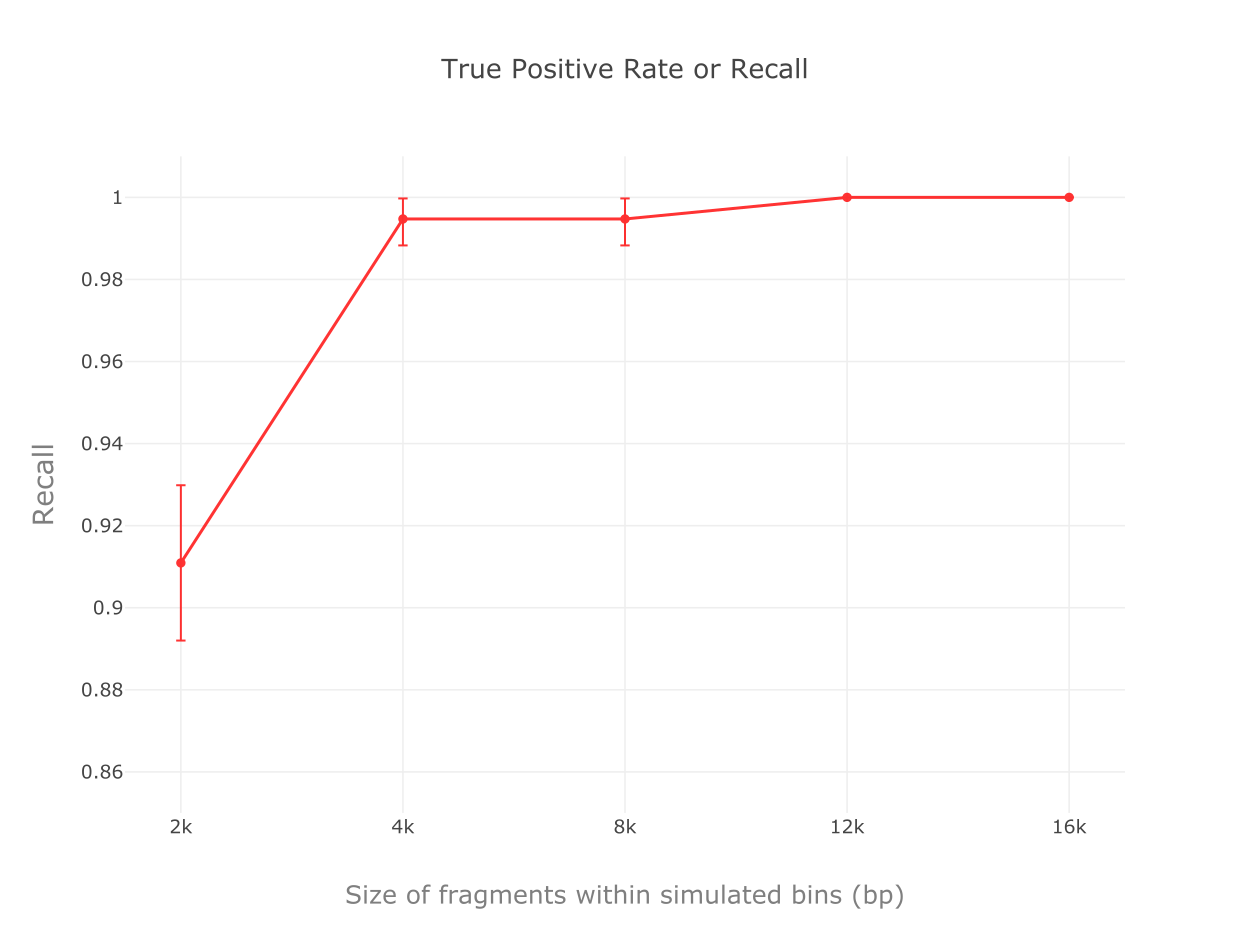
Recall rates for tests with different contig sizes.
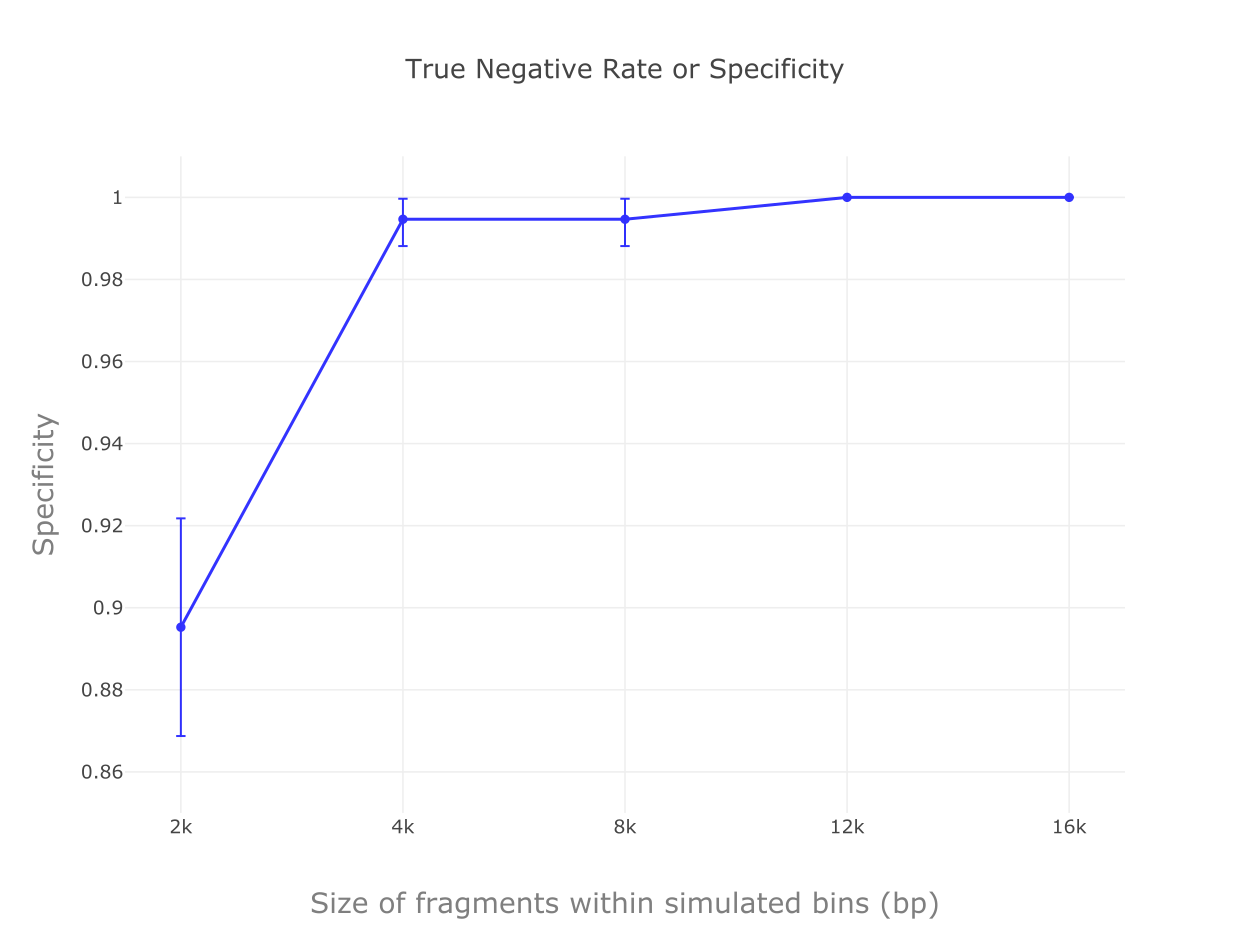
Specificity results for tests with different contig sizes.
Comparison with existing tools show that MARVEL’s superior performance for most assessed scenarios.